Now Reading: Advancing RNA Editing: Breakthrough with Enhanced Guide RNAs Mimicking ADAR Substrates
-
01
Advancing RNA Editing: Breakthrough with Enhanced Guide RNAs Mimicking ADAR Substrates
Advancing RNA Editing: Breakthrough with Enhanced Guide RNAs Mimicking ADAR Substrates
Quick Summary:
- Sequence data associated with the study is hosted on the National Centre for biotechnology Information under BioProject ID PRJNA1163502.
- Online resources available include TxDb.Hsapiens.UCSC.hg19.knownGene from Bioconductor and additional datasets linked in a referenced Nature article.
- MIRROR,a program version 1.0, is released on GitHub for free non-commercial use.
Indian Opinion Analysis:
The increasing availability of public genetic data repositories and open-source tools like MIRROR highlights India’s growing reliance on access to global scientific advancements. While such developments open avenues for collaboration and innovation in bioinformatics within India, there must also be attention towards local investment in genomic research infrastructures. Potential policy interventions ensuring ethical compliance and conducive pathways for biomedical applications could strengthen India’s position as an active contributor to genomic science rather than just a consumer or observer.
For further details, refer to: Source Link
Quick Summary:
- The article explores advancements in RNA editing technologies using CRISPR-Cas13 and ADAR enzymes.
- RNA editing techniques are discussed for their precision, efficiency, and adaptability to various applications.
- Recent studies highlight methods employing guide RNAs to target endogenous transcripts with high fidelity for therapeutic goals.
- innovations include chemically modified oligonucleotides and circular RNA guides tested on non-human primates.
Indian Opinion Analysis:
The advancements in RNA editing reflect promising opportunities for biomedical applications worldwide,including India.Given India’s strong presence in biotechnology research, these innovations could enhance local efforts towards gene therapy solutions for inherited diseases. Additionally, leveraging such technologies might potentially be instrumental in addressing public health challenges.However, regulatory oversight and ethical considerations about genome-editing tools need thorough examination as the country adapts these techniques into its healthcare system.
Read MoreIt appears that the input provided does not include a complete or coherent news story. As no substantive information about India or related topics has been presented in this raw text, I am unable to draft a meaningful Quick Summary and Indian Opinion Analysis based on the current input. If you could provide a specific and detailed news article about India, I’d be happy to assist in summarizing and analyzing it effectively!Quick Summary
- The article discusses various advancements in RNA editing technologies focusing on A-to-I RNA editing, which is facilitated by enzymes such as ADAR2.
- Several studies have explored the mechanisms behind RNA editing, including base-flipping and site selectivity by ADAR enzymes.
- Recent developments include designing small guide RNAs (sgRNAs) and algorithms like dsRID that identify potential double-stranded RNA (dsRNA) regions using sequencing data.
- Large-scale analyses revealed millions of genomic sites involved in A-to-I editing, mainly within human Alu repeats.
- The research aims at therapeutic applications, with implications for genetic disorders such as Alpha1-Antitrypsin Deficiency.
Indian Opinion Analysis
The advancements in RNA editing technology exemplify ongoing efforts toward precision medicine worldwide, inclusive of India’s participation in scientific innovation and collaboration opportunities. Given India’s robust pharmaceutical sector and ambition to become a biotechnology hub, these findings could open avenues for native research institutions to integrate cutting-edge genetic therapies into their portfolios.
Moreover, biomedical advancements hold promise for India’s sizable population affected by hereditary health conditions or rare diseases.leveraging these developments would require proactive engagement from Indian researchers to contribute towards global publications while addressing local health needs pragmatically.
For more: Read MoreQuick Summary
- A study led by Rui Zhang and Wenbing Yang involving scientists at Sun Yat-Sen University and RecoRNA Biotechnology focuses on RNA editing, using innovative methods to analyze genomic features efficiently.
- Various biotechnological tools, such as STAR aligner, ChIPseeker, and BEDTools, were employed for data analysis in the research.
- Financial support was provided by multiple Chinese national scientific programs and regional grants in guangzhou. The study also received support from Sun Yat-Sen University’s research funds.
- The work has led to filed patents related to RNA editing technologies. Some authors are shareholders in RecoRNA Biotechnology while others are employees there.
Indian Opinion Analysis
The research underscores the growing influence of cutting-edge biotechnology advancements within scientific hubs like Sun Yat-Sen University and private firms such as RecoRNA Biotechnology. while this particular study focuses on improving RNA editing techniques globally,its implications for India could center around fostering similar collaborative efforts between academia and industry partners to address pressing genetic research challenges efficiently. Investment in genomics is becoming more prominent; Indian institutions can take cues from China’s multi-channel public-private funding approach for scalable innovation in biotechnology fields.
Read more: http://www.nature.com/articles/s41587-025-02628-6/figures/7Quick Summary
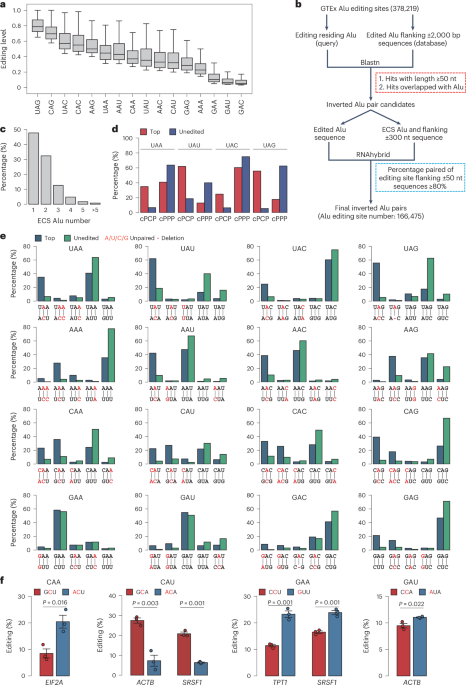
- The article discusses improvements in RNA base editing using specialized guide RNAs (gRNAs) called MIRROR gRNAs that mimic highly edited endogenous ADAR (adenosine deaminase acting on RNA) substrates.
- Extensive research is presented on editing sites, gRNA efficiencies, structural features impacting editing effectiveness, and the development of a high-throughput efficiency measurement system.
- Key advancements include prediction models using structural features for improved edits, frameworks for targeting specific gene regions like GAPDH, and optimizing chemical modifications of gRNAs.
- Experiments included targeted sequencing and screening systems in various biological models to evaluate reproducibility and accuracy.
Images from the study feature examples of gRNA designs, efficiency correlations across replicates, chemical modification testing results, secondary structure predictions of circular CLUSTER gRNAs targeting specific genes (TPT1, SRSF1), and quantitative analyses related to liver experiments with injected MIRROR gRNAs.
indian Opinion Analysis
This scientific advancement reflects growing sophistication in precision genetic technologies with potential global applications in medicine and research. For India specifically,this could bolster biotech industries focusing on therapeutic innovations-particularly RNA-based treatments-a field seeing increasing governmental emphasis under initiatives like ‘Make In India’ for biopharma sectors. The ability to target precise genome regions can aid domestic healthcare challenges related to inherited diseases or chronic conditions through personalized medicine approaches.
moreover, fostering collaboration between Indian researchers and international institutions could position the country as a meaningful contributor-or beneficiary-in breakthroughs like these by improving accessibility-wise affordability when applied locally. Biotech education may need refocusing around such cutting-edge topics to prepare future scientists for complex genomic solutions relevant to evolving healthcare demands.
Quick Summary
- Researchers have developed improved RNA base editing technology using guide RNAs that mimic highly edited endogenous ADAR substrates.
- The study, authored by Sun, Cao, Song et al.,has been published in Nature Biotechnology.
- The article was received on September 13, 2024, accepted on March 6, 2025, and published on April 3, 2025.
- The advancement is significant for RNA-based therapeutics and biotechnology applications.
Indian Opinion Analysis
This breakthrough in RNA base editing could hold relevance for India’s biotechnology sector as it continues to grow rapidly. India has seen increasing investment in genetic research and precision medicine in recent years; advances such as this may benefit future therapeutic developments targeted at India-specific health challenges like hereditary diseases or conditions prevalent among its diverse population. A strong focus on incorporating cutting-edge innovations like the one outlined here might enable better health outcomes and contribute to India’s emerging position as a global biotech hub.