Now Reading: Scientists Uncover Key Cause of Rising Methane Levels on Earth
-
01
Scientists Uncover Key Cause of Rising Methane Levels on Earth
Scientists Uncover Key Cause of Rising Methane Levels on Earth
Speedy Summary
- Methane, a potent greenhouse gas, is mainly produced by microbes (methanogens) in oxygen-free environments like wetlands, rice fields, and landfills.
- Tracking methane sources accurately remains challenging due to large uncertainties in its fluxes and isotope fingerprinting complexity.
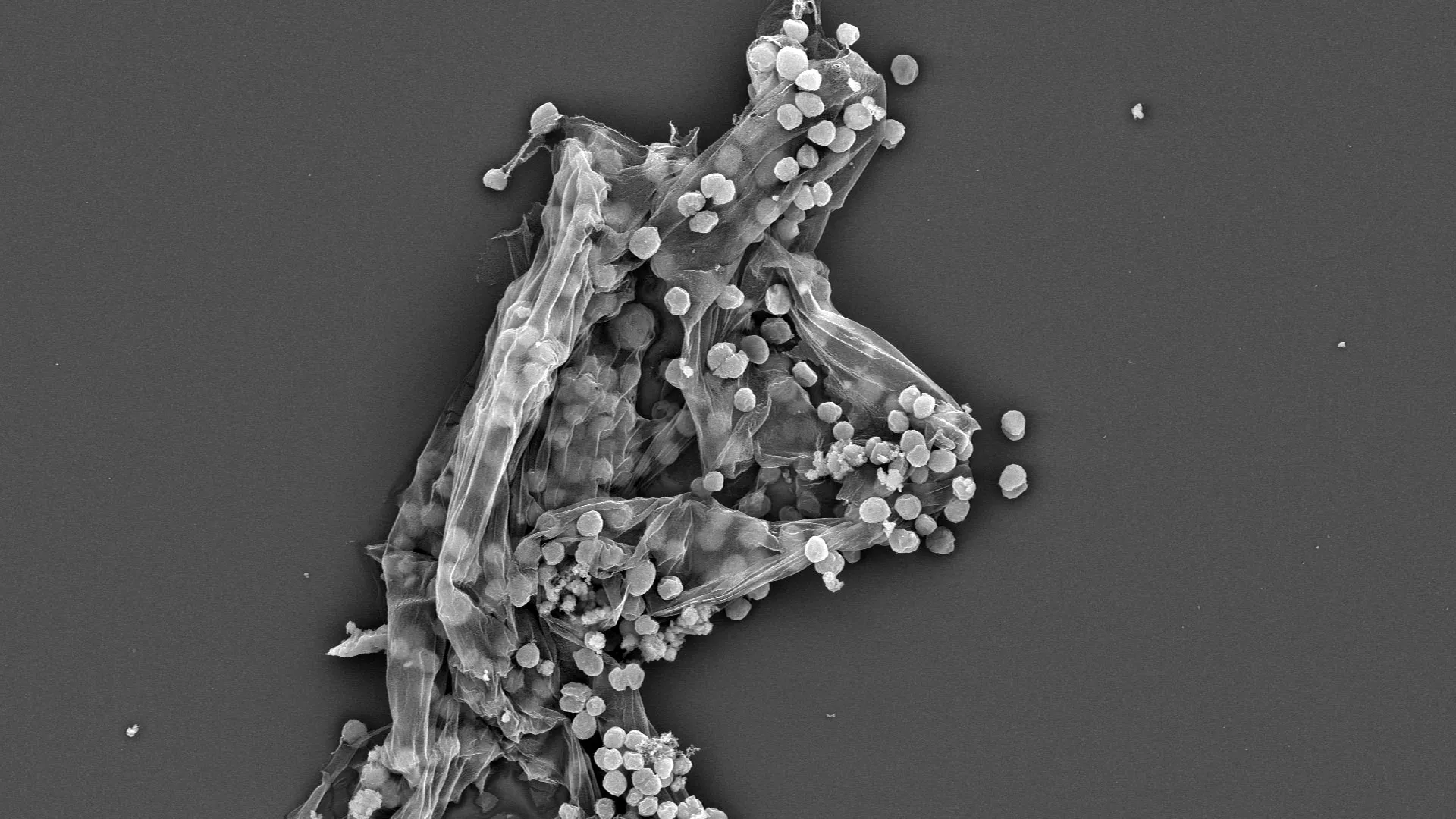
- A study by UC Berkeley researchers reveals how the key enzyme methyl-coenzyme M reductase (MCR) impacts methane’s isotopic composition through environmental or dietary conditions affecting microbes.
- The research used CRISPR gene editing to manipulate MCR activity in methanogens like Methanosarcina acetivorans, observing changes in isotopic fingerprints of methane under different conditions.
- They discovered that isotope exchange with water influences the hydrogen composition of microbial methane more than previously assumed. This indicates that acetate-consuming methanogens might have been underestimated as contributors to atmospheric methane.
- The findings propose integrating molecular biology with isotope geochemistry for better understanding and mitigating environmental issues related to methane production.
Indian Opinion analysis
The study sheds new light on the complexities of tracking and mitigating microbial contributions to global warming through advanced biotechnological approaches. For India-a notable producer of rice (a major source of wetland-related emissions)-this research could prove vital in refining national climate strategies.Improved knowledge about how specific environments contribute uniquely to methane emissions may enable more targeted interventions, especially for managing agricultural practices or landfill emissions.Collaboration between biological sciences and geochemistry offers novel opportunities globally; India’s scientific community stands poised to adapt similar interdisciplinary strategies tailored toward its unique environmental challenges.