Now Reading: Enhanced Microbial Identification in Metagenomic Data with SingleM and Sandpiper
-
01
Enhanced Microbial Identification in Metagenomic Data with SingleM and Sandpiper
Enhanced Microbial Identification in Metagenomic Data with SingleM and Sandpiper
Fast Summary:
- This article provides a detailed reference list on advancements in metagenomics tools and methodologies.
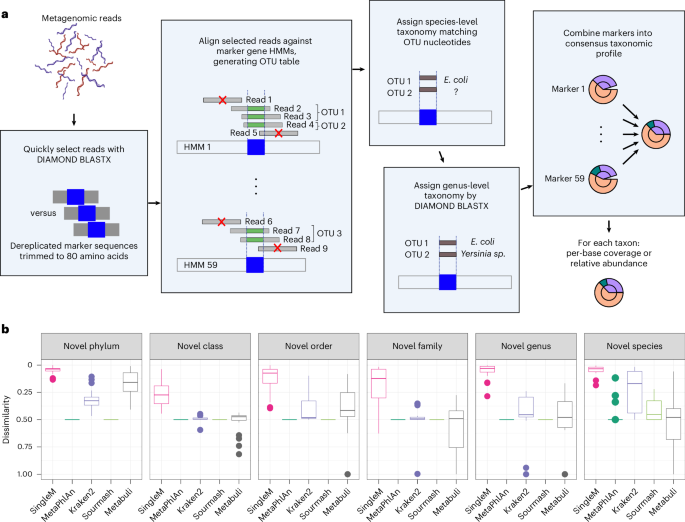
- It includes leading research contributions from international experts aimed at improving taxonomy profiling, false-positive removal, and genomic classification using diverse software such as Kaiju, DIAMOND, kraken 2, MetaPhlAn 4, and others.
- important concepts discussed include tree-of-life-scale protein alignments, cultivation-self-reliant genomes expansion (mOTUs), type IIB restriction sites targeting for false-positive removal, and crowdsourced benchmarking initiatives like sbv IMPROVER Microbiomics challenge.
Indian Opinion Analysis:
The refinement of metagenomic tools underscores the rising importance of microbiome studies globally. For India-a country with vast biodiversity spanning from forest ecosystems to urban landscapes-these advancements translate into opportunities for ecological monitoring and health sciences through DNA-level insights into microbial life. Such innovations could support analysis linked to agriculture sustainability or fight antimicrobial resistance prevalent in healthcare systems. However, fostering capacity-building efforts for implementation requires funding synchronicity between academia-industry partnerships within India’s bioinformatics sector while ensuring affordability accessibly aligns alongside ethical-genomic practices essential toward citizen-health privacy safeguards vital long-term scientific integration networks potential SE asian plains collaborative spectrum multi-purpose max utility meaningful scale program sustainable impact frameworks.”It seems that the input provided contains bibliographical references from scientific literature, rather than a news article about India. If you can provide the appropriate raw text of a news article specifically related to India, I’ll be able to craft a concise “Quick Summary” and “Indian Opinion Analysis” based on your needs. Let me know how I can assist further!Quick Summary
- The presented content lists references from research articles primarily focused on microbial communities, bioinformatics tools, microbiomes across various ecological environments, electron transfer in methane production processes, and metadata analysis techniques.
- Key subjects discussed include microbial resistome in migratory birds, freshwater species as reservoirs for pathogens like Vibrio cholerae, and the role of nanoparticles in biomedical applications.
- articles also explore the microbiomes of deep-sea fish, codfish intestinal ecosystems, large pelagic sharks with varied feeding ecologies, and correlations between macaque and human vaginal microbiomes for evolutionary studies.
- Bioinformatics advancements are highlighted through tools like Snakemake workflow engines or simulation methods used to analyze genomes/metagenomes.
for more: Referenced articles
Indian Opinion analysis
The selection of topics showcases India’s growing stake in leveraging biotechnology research to address both environmental challenges and medical innovations. With India being rich in biodiversity (coastal zones and freshwater resources), understanding resistome patterns among migratory species aligns well with safeguarding public health against emerging pathogens.Similarly, developments such as metagenomic profiling offer deep insights into unexplored microbiomes; this has applications for improving agricultural yields or marine ecology management within Indian waters.India’s recent focus on science-backed solutions via infrastructure upgrades could further benefit if these frameworks (like CheckM genome simulators) are localized or adapted efficiently into domestic research hubs. Sustainable implementation remains a logical next step toward translating scientific findings into actionable societal benefits across sectors like food security & ecosystem preservation.Quick Summary
- The text provided comprises reference citations from various studies,predominantly focusing on advancements in data analysis,graphics,microbial genome profiling tools,metagenomics utilities,and machine learning systems.
- Notable tools mentioned include R language for graphics and data analysis (1996), ggplot2 (2016), Metagenome profilers Struo2 (2021) and CheckM2 (2023), as well as scalable algorithms like Galah and SingleM for microbial genome quality assessment.
- There are links to Zenodo datasets, GitHub repositories for open-source code advancement in genomics/metagenomics research, PyPI archives for Python packages like SingleM, DockerHub containers for deployment testing tools such as SIngleM-installation.
indian Opinion Analysis
India’s scientific community can benefit greatly from these global advancements in bioinformatics technologies given its vibrant biotechnology sector. Tools such as CheckM2 or Metagenomic profilers could be applied across India’s diverse ecosystems to enhance genomic research capacities in agriculture and healthcare solutions-where India is striving to lead innovation globally. Emphasis should also be placed on nurturing local expertise around integrating open-source frameworks like those highlighted into India’s education system to bolster scalability efforts amidst rising population-driven resource needs.