Now Reading: Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor
-
01
Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor
Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor
Data availability
All deep sequencing data can be accessed at the National Center for Biotechnology Information (NCBI) BioProject under the accession code PRJNA1115824 (ref. 63). All whole-transcriptome sequencing data can be accessed in Gene Expression Omnibus under the accession codes GSE267726 (ref. 64), in the Genome Sequence Archive of the BIG Data Center at the Beijing Institute of Genomics, Chinese Academy of Science under the accession codes HRA007429 (ref. 65) and CRA016552 (ref. 66) and in the National Omics Data Encyclopedia under the accession code OEP00005360.
Human and mouse genome sequences were downloaded from University of California Santa Cruz (UCSC) Genome Browser (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz and https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/mm10.fa.gz). The single-nucleotide variants (SNVs) information for the GATK BaseRecalibrator of humans was downloaded from the Single-Nucleotide Polymorphism database (dbSNP) at the NCBI (ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b151_GRCh38p7/VCF/GATK/All_20180418.vcf.gz), and SNVs information for GATK BaseRecalibrator of the mouse was downloaded from the European Variation Archive (EVA) database at the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL–EBI) (ftp://ftp.ebi.ac.uk/pub/databases/eva/rs_releases/release_1/by_species/Mouse_10090/GRCm38.p4/GCA_000001635.6_current_ids.vcf.gz). Human and mouse repeat regions were downloaded from UCSC Genome Browser (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/database/rmsk.txt.gz and http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/rmsk.txt.gz). GATK VariantRecalibrator for humans, including multiple Variant Call Format (VCF) files, is available at https://console.cloud.google.com/storage/browser/_details/genomics-public-data/resources/broad/hg38/v0/hapmap_3.3.hg38.vcf.gz, https://console.cloud.google.com/storage/browser/_details/genomics-public-data/resources/broad/hg38/v0/1000G_omni2.5.hg38.vcf.gz, https://console.cloud.google.com/storage/browser/_details/genomics-public-data/resources/broad/hg38/v0/1000G_phase1.snps.high_confidence.hg38.vcf.gz, https://ftp.ncbi.nih.gov/snp/organisms/human_9606/VCF/00-All.vcf.gz and https://console.cloud.google.com/storage/browser/_details/genomics-public-data/resources/broad/hg38/v0/Mills_and_1000G_gold_standard.indels.hg38.vcf.gz. GATK VariantRecalibrator for mouse, including VCF file, is available at http://ftp.ebi.ac.uk/pub/databases/eva/rs_releases/release_1/by_species/Mouse_10090/GRCm38.p4/GCA_000001635.6_current_ids.vcf.gz. Files used to filter out the background variants for human are available at https://ftp.ncbi.nih.gov/snp/organisms/human_9606/VCF/00-All.vcf.gz and http://hgdownload.soe.ucsc.edu/goldenPath/hg38/database/rmsk.txt.gz. Files used to filter out the background variants for mice are available at http://ftp.ebi.ac.uk/pub/databases/eva/rs_releases/release_1/by_species/Mouse_10090/GRCm38.p4/GCA_000001635.6_current_ids.vcf.gz and http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/rmsk.txt.gz. Source data are provided with this paper.
Code availability
The computational pipeline for calculating frequencies of base substitution is available at GitHub (https://github.com/YangLab/CFBI). The computational pipeline of the RNA editing analysis pipeline to decode all 12 types of RNA editing events (RADAR) to perform transcriptome-wide off-target analysis is available at GitHub (https://github.com/YangLab/RADAR). The custom code for analyzing the efficiency and off-target effect of RtABE is available on GitHub at https://github.com/YangLab/RtABE_computational_analysis (ref. 67).
References
-
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR–Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
-
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
-
Yang, B., Yang, L. & Chen, J. Development and application of base editors. CRISPR J. 2, 91–104 (2019).
-
Yang, L. & Chen, J. A tale of two moieties: rapidly evolving CRISPR/Cas-based genome editing. Trends Biochem. Sci. 45, 874–888 (2020).
-
Li, G. et al. Gene editing and its applications in biomedicine. Sci. China Life Sci. 65, 660–700 (2022).
-
Li, X. et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat. Biotechnol. 36, 324–327 (2018).
-
Wang, J. Y. & Doudna, J. A. CRISPR technology: a decade of genome editing is only the beginning. Science 379, eadd8643 (2023).
-
Kim, J. S. & Chen, J. Base editing of organellar DNA with programmable deaminases. Nat. Rev. Mol. Cell Biol. 25, 34–45 (2024).
-
Khosravi, H. M. & Jantsch, M. F. Site-directed RNA editing: recent advances and open challenges. RNA Biol. 18, 41–50 (2021).
-
Pfeiffer, L. S. & Stafforst, T. Precision RNA base editing with engineered and endogenous effectors. Nat. Biotechnol. 41, 1526–1542 (2023).
-
Booth, B. J. et al. RNA editing: expanding the potential of RNA therapeutics. Mol. Ther. 31, 1533–1549 (2023).
-
Fu, Z. C., Gao, B. Q., Nan, F., Ma, X. K. & Yang, L. DEMINING: a deep learning model embedded framework to distinguish RNA editing from DNA mutations in RNA sequencing data. Genome Biol. 25, 258 (2024).
-
Merkle, T. et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37, 133–138 (2019).
-
Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019).
-
Reautschnig, P. et al. CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat. Biotechnol. 40, 759–768 (2022).
-
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
-
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
-
Reautschnig, P. et al. Precise in vivo RNA base editing with a wobble-enhanced circular CLUSTER guide RNA. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02313-0 (2024).
-
Riedmann, E. M., Schopoff, S., Hartner, J. C. & Jantsch, M. F. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA 14, 1110–1118 (2008).
-
Kuttan, A. & Bass, B. L. Mechanistic insights into editing-site specificity of ADARs. Proc. Natl Acad. Sci. USA 109, E3295–E3304 (2012).
-
Montiel-Gonzalez, M. F., Vallecillo-Viejo, I., Yudowski, G. A. & Rosenthal, J. J. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl Acad. Sci. USA 110, 18285–18290 (2013).
-
Vogel, P. et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018).
-
Kiran Kumar, K. D. et al. An improved SNAP-ADAR tool enables efficient RNA base editing to interfere with post-translational protein modification. Nat. Commun. 15, 6615 (2024).
-
Katrekar, D. et al. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 16, 239–242 (2019).
-
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
-
Kannan, S. et al. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 40, 194–197 (2022).
-
Liu, Y. et al. REPAIRx, a specific yet highly efficient programmable A > I RNA base editor. EMBO J. 39, e104748 (2020).
-
Xiao, Q. et al. Rescue of autosomal dominant hearing loss by in vivo delivery of mini dCas13X-derived RNA base editor. Sci. Transl. Med. 14, eabn0449 (2022).
-
Wang, L. et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat. Cell Biol. 23, 552–563 (2021).
-
Han, W. et al. Design and application of the transformer base editor in mammalian cells and mice. Nat. Protoc. 18, 3194–3228 (2023).
-
Han, W. et al. Base editing of the HBG promoter induces potent fetal hemoglobin expression with no detectable off-target mutations in human HSCs. Cell Stem Cell 30, 1624–1639 (2023).
-
Katrekar, D. et al. Comprehensive interrogation of the ADAR2 deaminase domain for engineering enhanced RNA editing activity and specificity. eLife 11, e75555 (2022).
-
Wang, Y. et al. ecRESCUE: a novel ecDHFR-regulated RESCUE system with reduced RNA off-targeting activity. Cell Commun. Signal. 19, 81 (2021).
-
Wang, X. et al. Cas12a base editors induce efficient and specific editing with low DNA damage response. Cell Rep. 31, 107723 (2020).
-
Liu, Z., Chen, S., Lai, L. & Li, Z. Inhibition of base editors with anti-deaminases derived from viruses. Nat. Commun. 13, 597 (2022).
-
Oakes, E., Anderson, A., Cohen-Gadol, A. & Hundley, H. A. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma. J. Biol. Chem. 292, 4326–4335 (2017).
-
Conticello, S. G., Harris, R. S. & Neuberger, M. S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13, 2009–2013 (2003).
-
Li, Y. L. et al. The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Nature 615, 728–733 (2023).
-
Salter, J. D., Bennett, R. P. & Smith, H. C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 41, 578–594 (2016).
-
Yang, B., Li, X., Lei, L. & Chen, J. APOBEC: from mutator to editor. J. Genet. Genomics 44, 423–437 (2017).
-
Pecori, R., Di Giorgio, S., Paulo Lorenzo, J. & Nina Papavasiliou, F. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat. Rev. Genet. 23, 505–518 (2022).
-
Xiang, J. et al. Nucleoside deaminases: the key players in base editing toolkit. Biophys. Rep. 9, 325–337 (2023).
-
Gray, D. C., Mahrus, S. & Wells, J. A. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 142, 637–646 (2010).
-
Szymczak-Workman, A. L., Vignali, K. M. & Vignali, D. A. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012, 199–204 (2012).
-
Matthews, M. M. et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat. Struct. Mol. Biol. 23, 426–433 (2016).
-
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
-
Pinto, Y., Cohen, H. Y. & Levanon, E. Y. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5 (2014).
-
Wang, D. et al. Characterization of an MPS I-H knock-in mouse that carries a nonsense mutation analogous to the human IDUA-W402X mutation. Mol. Genet. Metab. 99, 62–71 (2010).
-
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
-
Yang, L., Yang, B. & Chen, J. One prime for all editing. Cell 179, 1448–1450 (2019).
-
Chen, P. J. et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652 (2021).
-
Li, X. et al. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat. Commun. 13, 1669 (2022).
-
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
-
Lykke-Andersen, S. & Jensen, T. H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 (2015).
-
Yi, Z. et al. Utilizing AAV-mediated LEAPER 2.0 for programmable RNA editing in non-human primates and nonsense mutation correction in humanized Hurler syndrome mice. Genome Biol. 24, 243 (2023).
-
Monian, P. et al. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol. 40, 1093–1102 (2022).
-
Erion, D. M., Liu, L. Y., Brown, C. R., Rennard, S. & Farah, H. Editing approaches to treat α-1 antitrypsin deficiency. Chest 167, 444–452 (2024).
-
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
-
Rehmsmeier, M., Steffen, P., Hochsmann, M. & Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 (2004).
-
Wang, L. et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 27, 1289–1292 (2017).
-
Lei, L. et al. APOBEC3 induces mutations during repair of CRISPR–Cas9-generated DNA breaks. Nat. Struct. Mol. Biol. 25, 45–52 (2018).
-
Wang, X. et al. Efficient base editing in methylated regions with a human APOBEC3A–Cas9 fusion. Nat. Biotechnol. 36, 946–949 (2018).
-
Li, G. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1115824 (2025).
-
Li, G. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE267726 (2025).
-
Li, G. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor. https://ngdc.cncb.ac.cn/gsa-human/browse/HRA007429 (2025).
-
Li, G. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor. https://ngdc.cncb.ac.cn/gsa/browse/CRA016552 (2025).
-
Li, G. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor. GitHub https://github.com/YangLab/RtABE_computational_analysis (2025).
Acknowledgements
This work is supported by the National Key Research and Development Program of China (2023YFC3403402 to J.C., 2024YFC3405902 to L.Y., 2024YFA1803301 to J.C. and 2023ZD0500501 to B.Y.), the National Natural Science Foundation of China (32371514 to J.C., 32430018 to L.Y. and 32070170 to B.Y.), the Ministry of Agriculture and Rural Affairs of the People’s Republic of China (NK2022010207 to J.C.), the Shanghai Municipal Science and Technology Commission (23JS1400300 and 23DX1900102 to L.Y. and 23ZR1442500 to B.Y.) and the Program of Shanghai Academic/Technology Research Leader (23XD1422500 to J.C.). We thank Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine, ShanghaiTech University, Molecular and Cell Biology Core Facility, School of Life Science and Technology, ShanghaiTech University, Animal Core Faculty, ShanghaiTech University, the Core Facility of Shanghai Medical College, Fudan University and the Medical Science Data Center in Shanghai Medical College of Fudan University for the technical support.
Ethics declarations
Competing interests
J.C., L.Y. and B.Y. are scientific cofounders of CorrectSequence Therapeutics, a company that uses gene-editing technologies, and the members of its scientific advisory board. J.C., L.Y., B.Y., G.L. and G.C. have filed patent applications relating to the published work through ShanghaiTech University. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Hui Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison of previously published RNA base editing systems.
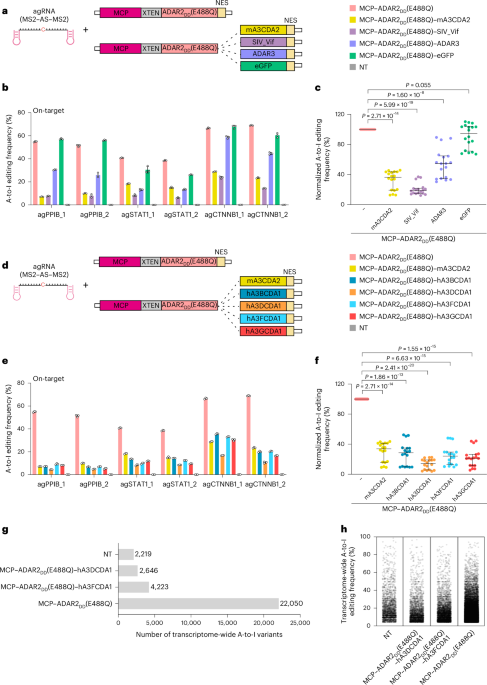
a, The expression of ADAR1 and ADAR2 in 293FT, HeLa and U2OS cells. ADAR1 and ADAR2 mRNA levels were determined via RT–qPCR and normalized to the level of GAPDH. b,c, Comparison of the on-target A-to-I editing efficiencies induced by the indicated RNA base editing systems in 293FT cells. d, Numbers of transcriptome-wide A-to-I variants in 293FT cells transfected with the MCP–ADAR2DD(E488Q) RNA editing system against the CTNNB1_1 target site. NT, nontransfected control. e, Manhattan plot of transcriptome-wide A-to-I editing frequencies in 293FT cells transfected with the MCP–ADAR2DD(E488Q) RNA editing system against the CTNNB1_1 target site. For a–c, the data are presented as the mean ± s.d. from three independent experiments.
Extended Data Fig. 2 Effects of inhibitors or RNA-binding protein on ADAR2DD.
a, Western blot analysis of the expression of MCP-ADAR2DD(E488Q) and the MCP-ADAR2DD(E488Q) fused to potential inhibitor proteins in 293FT cells. GAPDH was used as the internal control. The blots for anti-MCP and anti-GAPDH were performed with the same samples but run in different gels. b, Schematic of the fusion of ADAR2DD(E488Q) to different RNA-binding proteins at N-terminus, without or with the fusion of A3DADI at C-terminus. XTEN, linking peptides. c, On-target A-to-I editing efficiencies induced by the fusion proteins shown in b with the gRNAs against the CTNNB1_2 site in 293FT cells. The data are from one experiment. d, Numbers of transcriptome-wide A-to-I variants and Manhattan plot of transcriptome-wide A-to-I editing frequencies in 293FT cells transfected with the indicated proteins and the guide RNA targeting CTNNB1_2 site. e, Western blot analysis of the expression of ADAR2DD(E488Q)–Flag and the ADAR2DD(E488Q)–Flag fused to MCP or eGFP at N-terminus. GAPDH was used as the internal control. The blots for anti-Flag and anti-GAPDH were performed with the same samples but run in different gels. For a and e, the experiments were repeated three times independently with similar results, and the results from one representative experiment are shown.
Extended Data Fig. 3 Cleavage of A3DADI triggered by different versions of TEV proteases and guide RNAs.
a, Western blot analysis of the cleavage of MCP-ADAR2DD(E488Q)-TS-A3DADI in the presence of intact TEVp or two versions of split TEVp in 293FT cells. GAPDH was used as the internal control. The blots for anti-Flag and anti-GAPDH were performed with the same samples but run in different gels. b, Western blot analysis of the cleavage of MCP-ADAR2DD(E488Q)-TS-A3DADI in the presence of different versions of guide RNAs targeting CTNNB1 in 293FT cells. GAPDH was used as the internal control. The blots for anti-Flag and anti-GAPDH were performed with the same samples but run in different gels. For a and b, the experiments were repeated three times independently with similar results, and the results from one representative experiment are shown.
Extended Data Fig. 4 Off-target editing in exon regions and minimal free energy between guide RNA and the identified off-target sites.
a, Numbers of A-to-I variants and Manhattan plot of A-to-I editing frequencies in exons induced by the indicated RNA base editing systems targeting CTNNB1_1 in 293FT cells. CDS, coding sequence; UTR, untranslated region. b,c, Minimal free energy of the AS region of eagCTNNB1_1 and the off-target sites identified for RtABE_V1 (b) and RtABE_V2 (c) in a by RNAhybrid.
Extended Data Fig. 5 Analyses of editing index and conserved ADAR target sites.
a, A-to-G RNA editing index in coding sequences for nontransfected (NT), MCP-ADAR2DD(E488Q)-transfected, RtABE_V1-transfected and RtABE_V2-transfected 293FT cells with the guide RNAs targeting CTNNB1_1, PPIA_5 and STAT1_3. b, Numbers of A-to-I variants and Manhattan plot of A-to-I editing frequencies in the transcriptomes in nontransfected 293FT cells, hADAR2-transfected 293FT cells and 293FT cells transfected with RtABE_V1 or RtABE_V2 targeting CTNNB1_1. The conserved ADAR target sites are shown in purple.
Extended Data Fig. 6 Characterization of RtABE in other cells and its effect on gene expression.
a,b, On-target A-to-I editing efficiencies induced by RtABE_V1 and RtABE_V2 at four indicated target sites in HeLa (a) and U2OS (b) cells. The data are presented as the mean ± s.d. from three independent experiments. c, Heatmap of Pearson correlations of gene expression profiles (fragments per kilobase of transcript per million mapped fragments) among samples from nontransfected 293FT cells (NT) and 293FT cells transfected with eGFP or RtABEs targeting CTNNB1_1.
Extended Data Fig. 7 Effects of AS region length and target adenosine position on editing efficiency and minimization of bystander editing via the depletion of a bystander-opposite base in eagRNA.
a, Schematics of the eagRNAs targeting the CTNNB1 transcript with different lengths of the AS region or with the target adenosine at different positions. b, Heatmap of the A-to-I editing frequency at each adenosine site in the target region of the CTNNB1 transcript. The red boxes demonstrate the adenosines covered by the AS regions of the indicated eagRNAs shown in a. c, Schematics of eagRNAs targeting the CTNNB1 transcript with the deletion of bystander-opposite base in the AS region to reduce bystander editing. d, Heatmap of the A-to-I editing frequencies induced by the eagRNAs shown in c at the indicated adenosine sites.
Extended Data Fig. 8 Generation of ADAR1−/− 293FT cells.
a, Sanger sequencing was used to determine the genotype of the ADAR1−/− 293FT cells generated with Cas9 nuclease. b, Western blot analysis of ADAR1 protein expression in wild-type 293FT and ADAR1−/− 293FT cells. GAPDH was used as the internal control. The blots for anti-ADAR1 and anti-GAPDH were performed with the same samples but run in different gels. The experiments were repeated three times independently with similar results, and the results from one representative experiment are shown.
Extended Data Fig. 9 In vitro test of RtABE targeting the W392X mutation of mIdua.
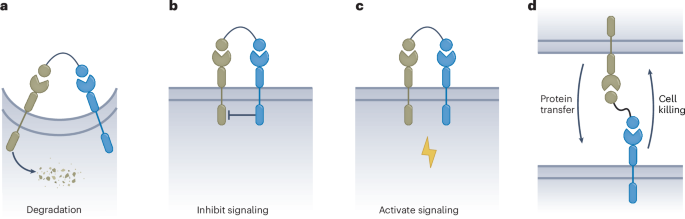
a, Schematics illustrating the generation of N2A cells harboring the mIduaW392X pathogenic mutation and its reversion by RNA editing. b, Sanger sequencing was used to determine the genotype of the IDUA–W392X N2A cells generated with prime editor 5. c, On-target A-to-I editing frequencies induced by RtABE_V1 and RtABE_V2 in IDUA-W392X N2A cells. d, Catalytic activities of α-l-iduronidase in wild-type (WT) N2A cells and IDUA-W392X N2A cells transfected with RtABE_V1 or RtABE_V2. The average activity of WT N2A was set as 100%. For c and d, the data are presented as the mean ± s.d. from three independent experiments. e, Schematic of the AAV packaging of the MCP–ADAR2DD(E488Q) RNA editing system. f, Schematics illustrating the potential working mechanism of RtABE. Newly transcribed eagRNAs may form secondary structures through intramolecular base pairing. At the on-target site, MS2 and BoxB aptamers can form and recruit inactive ADAR2DD and TEVc, respectively. The free diffusion of TEVn can reconstitute an intact TEVp at the on-target site, which leads to the cleavage of A3DADI. Finally, active ADAR2DD can induce specific and efficient A-to-I editing at the on-target site.
Extended Data Fig. 10 Cleavage of A3DADI when the target transcript was knocked down.
a, Examination of knockdown effect of the siRNA targeting CTNNB1 (siRNA_CTNNB1) with RT–qPCR. The data are from one experiment. b, Western blot analysis of the cleavage of MCP-ADAR2DD(E488Q)-TS-A3DADI when the target transcript was knocked down. The samples without eagRNA or with the eGFP-targeting eagRNA were as controls. GAPDH was used as the internal control. The blots for anti-Flag and anti-GAPDH were performed with the same samples but run in different gels. The experiments were repeated three times independently with similar results and the results from one representative experiment are shown.
Supplementary information
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, G., Chen, G., Yuan, GH. et al. Specific and efficient RNA A-to-I editing through cleavage of an ADAR inhibitor.
Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02591-2
-
Received:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41587-025-02591-2