Now Reading: The Protein Design Archive (PDA): insights from 40 years of protein design
-
01
The Protein Design Archive (PDA): insights from 40 years of protein design
The Protein Design Archive (PDA): insights from 40 years of protein design
While natural proteins are incredibly diverse biomacromolecules, they present only a subset of the sequence and structural space that is chemically possible1. The field of protein design has the potential to illuminate the unexplored regions of this space, to deepen our understanding of sequence–structure–function relationships, and to enable the development of novel proteins that could be used to address societal challenges.
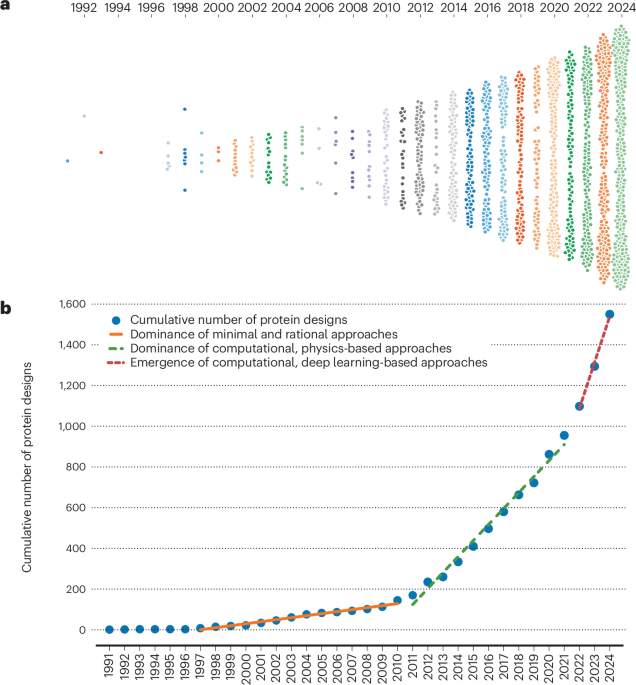
Protein design has evolved dramatically over the past 40 years from rational design to contemporary data-driven approaches. The field has now reached milestones such as the design of small-molecule2 and protein3 binders, entirely novel folds4,5,6, and has been recognized with a Nobel Prize. However, it can be difficult to identify current challenges and opportunities within the field as no easily searchable resource provides an overview of proteins that have been designed to date. Such a resource would improve our ability to learn from previous efforts and guide the development of future design methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
24,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
195,33 € per year
only 16,28 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Data availability
The PDA is available without registration at https://pragmaticproteindesign.bio.ed.ac.uk/pda/. Source data are provided with this paper.
Code availability
Code supporting PDA data collection and processing is available on GitHub: https://github.com/wells-wood-research/chronowska-stam-wood-2024-protein-design-archive. This includes files detailing manual curation of the PDA dataset, such as csv files listing proteins that were manually included and excluded, and how information was manually corrected. Code used to build the PDA website is available on GitHub: https://github.com/wells-wood-research/protein-design-archive.
References
-
Baker, D. Protein Sci. 28, 678–683 (2019).
-
Lu, L. et al. Science 384, 106–112 (2024).
-
Gainza, P. et al. Nature 617, 176–184 (2023).
-
Kuhlman, B. et al. Science 302, 1364–1368 (2003).
-
Thomson, A. R. et al. Science 346, 485–488 (2014).
-
Anishchenko, I. Nature 600, 547–552 (2021).
-
Burley, S. K. et al. Nucleic Acids Res. 49, D437–D451 (2021). (D1).
-
Leaver-Fay, A. et al. Methods Enzymol. 487, 545–574 (2011).
-
Chaudhury, S., Lyskov, S. & Gray, J. J. Bioinformatics 26, 689–691 (2010).
-
Stam, M. J. & Wood, C. W. Protein Eng. Des. Sel. 34, gzab029 (2021).
-
Steinegger, M. & Söding, J. Nat. Biotechnol. 35, 1026–1028 (2017).
-
van Kempen, M. Nat. Biotechnol. 42, 243–246 (2024).
-
Huang, P.-S., Boyken, S. E. & Baker, D. Nature 537, 320–327 (2016).
-
Woolfson, D. N. J. Mol. Biol. 433, 167160 (2021).
-
Kortemme, T. Cell 187, 526–544 (2024).
Acknowledgements
We thank the members of the Wells Wood Research Group for testing and feedback on the PDA website. M.C. is supported by a PhD studentship from the UK Research and Innovation-funded EastBio Doctoral Training Partnership programme. M.J.S., C.W.W. and D.N.W. are supported by a UKRI Biotechnology and Biological Sciences Research Council Strategic Longer and Larger award (BB/X003027/1).
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4, Tables 1–4, Discussion, Methods, Software Versions, Front-end Requirements, Back-end Requirements, References
Source data
Source Data Fig. 1
The Protein Design Archive dataset as of 1 January 2025
Source Data Fig. 2
Structure-based metrics generated with DE-STRESS for proteins found in the Protein Design Archive database as of 1 August 2024 and the Research Collaboratory for Structural Bioinformatics Protein Data Bank as of 1 June 2024
Rights and permissions
About this article
Cite this article
Chronowska, M., Stam, M.J., Woolfson, D.N. et al. The Protein Design Archive (PDA): insights from 40 years of protein design.
Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02607-x
-
Published:
-
DOI: https://doi.org/10.1038/s41587-025-02607-x